Genetic Networks
How do the interactions among genes influence health and development?
Our genes determine all sorts of things about us, including our predisposition to many diseases. But there are only a small number that we understand well enough to know how a genetic change is propagated through the network of molecular and cellular interactions to ultimately yield a human disease.
If we can get a better understanding of how these interactions work, and of how the effects of genetic perturbation propagate through the system, we will be better able to identify the root causes of many complex genetic diseases like autism, asthma, Alzheimer’s and many cancers.
The Genetic Networks program at CIFAR is charting genetic and molecular interactions to understand how biological systems work and how they fail. The aim is to map complete networks of genetic and molecular interactions, and to use them to decipher the rules of how genes influence one another, what environmental factors alter those interactions, and how the impact of genetic change is propagated through biological systems.
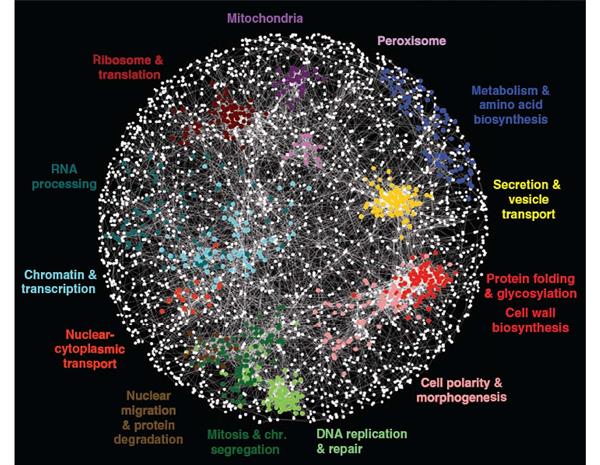
The Genetic Networks Program brings together geneticists with molecular and computational biologists, who work on a wide variety of species, from yeasts, fruit flies, worms and mice to humans. Because evolution has preserved many genes and genetic interactions over millions of years – for instance, humans and yeast share about 40 percent of their genes – research into the simpler organisms can shed light on humans. This broadly integrated investigation of genetic networks is unique in the world and has helped to recruit eminent researchers and give them the opportunity to connect and collaborate with leading researchers from other countries. Their work is uncovering how networks of molecular interactions mediate the effects of combinations of genetic perturbations, yielding maps from personal genomes to states of health and disease. The multidisciplinary Genetic Networks program has benefited greatly from the cutting-edge work of other CIFAR programs. For example, program researchers have used deep learning — a machine learning technique pioneered by CIFAR fellows in the Learning in Machines & Brainsprogram (formerly known as Neural Computation & Adaptive Perception) — to better predict how genetic change affects human disease.
A recent explosion in gene sequencing technologies has identified a massive catalogue of genes. However, we still need to understand how the information that’s encoded in genes, and the vast number of interactions between them, translates into the specific traits and characteristics that are unique to every one of us. By developing methods that predict the direct outcomes, including disease, from an individual’s complex genetic makeup, the program is helping to lay the groundwork that will lead to medicine that is personalized according to each individual’s genome. Researchers have made progress deciphering the causes of diseases from single genes, like cystic fibrosis and Huntington’s disease. But what’s needed now is a better understanding of how multiple genes mutate, combine, and alter networks of molecular and cellular interactions to cause more complex diseases. Even though there are hundreds of human gene variants, many lead to disease only in specific combinations.
Program members, individually and collaboratively, have made major progress in mapping genetic interactions. Partial network maps now exist for several model organisms, including two different yeast species, and the nematode worm. Work on genetic network mapping in cultured human cells is also underway. Program members have tested how genetic interactions are conserved between species – a key question for applying knowledge of model systems to more complex organisms, including humans. New genetic interaction maps and conservation studies have become starting points for program researchers to address broad evolutionary questions and better understand the genetic basis of many human diseases. Brendan Frey, a senior fellow in both NCAP and Genetic Networks, used a new computational technique to reveal tens of thousands of genetic variants that provide insight into the genetic basis of spinal muscular atrophy, hereditary nonpolyposis colorectal cancer and autism spectrum disorder.
SELECTED PAPERS
Davierwala, A.P. et al., “The synthetic genetic interaction spectrum of essential genes,” Nature Genetics 37 (2005): 1147-52. ABSTRACT
Redon, R. et al., “Global variation in copy number in the human genome,” Nature 444 (2006): 444-454. ABSTRACT
Levy, S. et al., “The Diploid Genome Sequence of an Individual Human,” PLoS Biology 5, 10 (2007): e254. ABSTRACT
Barash Y. et al, “Deciphering the splicing code,” Nature 465, 7294 (2010): 53–59. ABSTRACT
Founded
2005
Renewal Dates
2010, 2016
Interdisciplinary Collaboration
Genetics
Biochemistry
Molecular biology
Computational biology
Bioinformatics
Cell, evolutionary and systems biology
Pathology
Immunology
Biotechnology


















